

Next: About this document ...
Tik-61,181: Bioinformatics Kaski,Mannila,Nikkilä
Exercises 1, Autumn 2000
In essay type answers try to answer briefly and concentrate on
relevant issues. It is assumed that the assistant will need only
reasonable amount of time when decoding the answers. If the opposite
occurs, it may affect your grade.
The maximum length of an answer to one question is one page. If you make
some proofs or simulations they can be included as appendixes. Every
answer will be evaluated with the scale {0:failed, 3:accepted,
5:accepted with distinction}. In order to pass the course you need at
least grade 3 for 60% of the exercises. In order to pass with
distinction you need to return 95% of the exercises, and to get the
grade 5 for most of them.
The deadline of the exercises is 31.1.2001.
- a)
- Explain in detail how you would align the following
sequences with pair HMM:s and with non-probabilistic methods:
AAAG
ACG
(DEKM ch.2 and ch.4, S&M ch.3)
- b)
- Analyze briefly the relation of the probabilistic approach and
the non-probabilistic approach for sequence alignment.
(DEKM ch.2 and
ch.4, S&M ch.3)
- c)
- Give an example of the possible uses of the MC-model and HMMs in
bioinformatics. Outline briefly the main differences between the two models.
(DEKM ch.3)
- d)
- Relax. You are buying a drink from the notorious crazy coke
machine while absent-mindedly solving some exercises. The machine can be
in either of two states: cola preferring state (CP) and mineral
water (WP) preferring state. When you put in a coin, the output of the
machine can be described with the following probability matrix:
| |
cola |
mineral water |
lemonade |
| CP |
0.6 |
0.1 |
0.3 |
| WP |
0.1 |
0.7 |
0.2 |
Draw the state model graph with transition probabilities
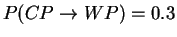 and
and
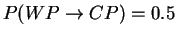 and calculate the probability of seeing the output
sequence {cola,lemonade} if the machine always starts off in the WP
state. What kind of behaviour would make this HMM to a (visible)
Markov model?
and calculate the probability of seeing the output
sequence {cola,lemonade} if the machine always starts off in the WP
state. What kind of behaviour would make this HMM to a (visible)
Markov model?
(for example DEKM ch.3)
- e)
- Do the exercise 3.3 in DEKM and explain its result briefly
in the bioinformatics framework.
- f)
- Compare briefly the FSAs and pairwise HMMs in searching.
(DEKM ch.4)
- g)
- Analyze the relation of the profile HMMs, HMMs, and PSSMs.
- h)
- Exercise 6.1 in DEKM.
- i)
- Explain briefly the pros, cons, and the most useful
applications of the different progressive alignment methods and
profile HMMs in multiple alignment.
(DEKM ch.6)
- j)
- Explain the reasons why there are methods for fragment
assembly and physical mapping.
(S&M: ch.4 and ch.5)
- k)
- Exercise 1 in ch.4 in S&M.
- l)
- Exercise 5 in ch.5 in S&M.
- m)
- Exercise 6 in ch.6 in S&M.
- n)
- Exercise 8.2 in DEKM. Explain also the concept of multiplicativity here.
- o)
- Exercise 8.7 in DEKM


Next: About this document ...
Janne Nikkila
2000-11-06